 |
Finn Årup Nielsen
We describe the ``Brede'' neuroinformatics database that provides data for novel information retrieval techniques and automated meta-analyses.
The database is inspired by the hierarchical structure of BrainMap [1] with scientific articles (``bib'' structures) on the highest level containing one or more experiments (``exp'' structure'', corresponding to a contrast in general linear model analyses), these in turn comprising one or more locations (``loc'' structures). The information on the ``bib'' level (author, title, ...) is setup automatically from PubMed while the rest of the information is entered manually in a Matlab graphical user interface. On the ``loc'' level this includes the 3D stereotactic coordinates in either Talairach or MNI space, the brain area (functional, anatomical or cytoarchitectonic area) and magnitude values such as Z-score and P-value. On the ``exp'' level information such as modality, scanner and behavioral domain are recorded with ``external components'' (such as ``face recognition'' or ``kinetic boundaries'') organized in a directed graph and marked up with Medical Subject Headings (MeSH) where possible.
Items in the database are identified with unique numbers and the type of identifier is given a unique string, e.g., ``WOBIB: 27'' for an Epstein and Kanwisher paper. This will allow Internet search engine to identify the phrase. For storing the data we employ a simple XML format that we denote ``poor-man's'' XML (pXML) with no attributes and no empty tags. The database presently consists of data constructed from 40 scientific articles, containing 134 experiments and 882 locations
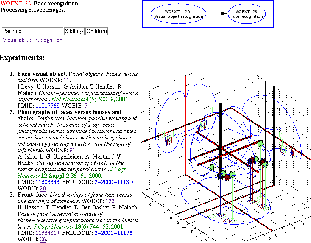
Static web-pages are generated from the ``exp'' and ``bib'' structures with Corner Cube visualization [2] as PNG and VRML files and hyperlinks to PubMed and fMRIDC [3]. The locations for each ``exp'' and ``bib'' structure are voxelized to a volume by convolving each location with a Gaussian kernel [4]. The combined set of volumes are converted to matrices and ``automated'' multivariate analyses are performed such as singular value decomposition (SVD) and independent component analysis (ICA). Sorted lists with related volumes are found for each individual volume as well as with respect to the SVD eigenimages and the results of the ICA. Ad hoc search can obtain the closest locations to a user-specified coordinate or the closest experiments to a user-specified set of locations.
The database is distributed as part of the Brede neuroinformatics toolbox (hendrix.imm.dtu.dk/software/brede/, [5]) which also provides the functions to manipulate and analyze the data. It is also available directly on the web (hendrix.imm.dtu.dk/services/jerne/).
EU Project MAPAWAMO (QLG3-CT-2000-300161).